Recent research into the gastrointestinal microbiome has confirmed the microbial inhabitants of the gastrointestinal tract, and their vast scope of metabolic activities, are at the very core of health and many disease processes. Microbiology testing should be optimized to address the abundance of all bacterial species present in a stool specimen.
This blog aims to summarize and weigh up the two current testing methodologies used to study gastrointestinal flora.
Proteomic Analysis (MaLDI-TOF)
This method is based on the culture method which is considered the industry gold standard as it enables identification of living metabolically active micro-organisms.
MaLDI-TOF technology identifies the micro-organisms by Proteomic Analysis which identifies the proteins in cells, tissues or organisms. Microorganisms have unique ribosomal protein ‘fingerprints’.
MaLDI-TOF stands for: Matrix- assisted Laser Desorption/Ionization Time of Flight Mass Spectroscopy
- MaLDI-TOF allows identification of over 7,000 micro-organisms! Traditional culture methods identify around 400.
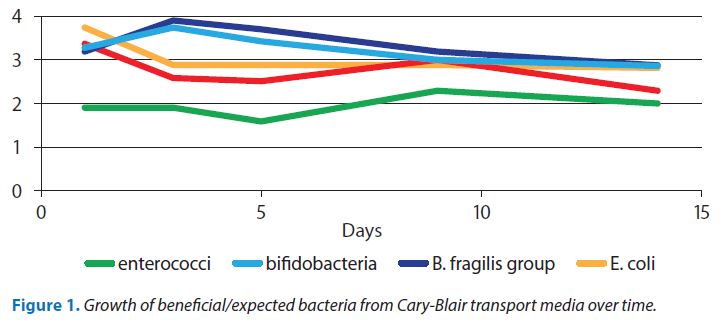
- The stool sample is collected into a preservative solution called ‘Clary Blair Transport Medium’ to prevent continued growth of flora whilst keeping the micro-organisms alive. The medium is buffered to prevent shifts in pH during transport, and contains a reducing agent designed to markedly decrease oxygen levels to favour the facultative and strict anaerobic species known to inhabit the GI tract. When properly collected and shipped, beneficial and pathogenic species may be successfully recovered quantitatively from the transport media for up to 14 days. showing proof of this concept – figure 1
- MaLDI-TOF has been used to study a host of macromolecules from a wide variety of biological matrices, and similar methods of detection have been used by NASA on the Mars Curiosity Rover in an effort to determine if the planet had been hospitable for microbial inhabitance.
- The MaLDI-TOF approach has an impressive capability to differentiate closely related species from a genus such as Bacillus.
- The results of an international, multicentre trial to evaluate the reproducibility of a commercially available MaLDI-TOF MS platform were presented at the Congress of Clinical Microbiology and Infectious Diseases in 2009. Sixty blind-coded bacterial specimens were sent to eight different international laboratories. A concordance rate of 98.8% was achieved among labs for the 480 specimens. Not only were the samples correctly identified across laboratories, but the reliability of identification was also very high as 97.3% of all log score values were > 2.
- Considering accuracy, inter laboratory reproducibility, cost and the rapidity of identification the proteonomic approach (MALDI-TOF MS) is best suited for routine comprehensive stool analysis.

The 5 steps in the MaLDI-TOF MS identification process:
- onization of proteins from a pure isolate (with a lazer)
- Separation of the vaporized, ionized proteins that have different masses (through the mass spectrometer)
- Detection of the number of the different ionized proteins
- Collection of the data
- Comparison for identification to a huge data base
Identification by PCR (DNA) Probes
Polymerase chain reaction (PCR) is a relatively old biochemical technology that is used in molecular biology to amplify segments of DNA. After amplification several different means of analysis have been applied in order to identify the preselected target segment of DNA.
- PCR probes have proven useful for clinical microbiology in hospital settings for rapid (<3 hours) identification of causative bacteria associated with serious diarrheal diseases. The speed and accuracy of the PCR platforms is due to the fact the bacteria do not have to be cultured.
- Research demonstrated that the PCR platform is very sensitive and precise for quantitative detection of human pathogens from fecal samples.
- PCR is also useful for characterization of abundant microbial communities clustered into enterotypes that appear to be associated with health and specific diseases.
Disadvantages of PCR (DNA) Probes
- PCR probes are only able to identify the specific item they were designed to discover (not broad spectrum). A primer is required for identification of every genus (let alone species) Therefore, a reliable PCR based evaluation of microorganisms in a stool specimen that is even 1/10th as comprehensive as that of the culture/MaLDI- TOF approach would be cost prohibitive.
- Detection limit – The ability for a DNA sequence probe to fit lock and key with another DNA means it has to have access so emulsion technologies are used; the surface area is spread to access more of the sample, however with this technology thousands and thousands of the organism are required to be picked up.
- PCR probes cannot differentiate between live bacteria and fragments of dead bacteria (e.g: from food).
- PCR-based platforms have significant limitations in the context of clinical medicine, especially for Complementary and Integrative medicine (CAM) in a non-acute care setting. CAM-oriented clinicians are interested in the composition and quantity of gastrointestinal microbiota, even in the absence of entero pathogens. It is the cause of changes in colonic metabolism that can be associated with symptoms of dysbiosis and various diseases.
Summary
MALDI-TOF/culture approach provides as much information as possible about the vast array of microorganisms that are present in a patients’ stool specimen whereas PCR provides information on a handful of microorganisms. In essence PCR-based methods are better suited to address the question “is it there” rather than “what is there?”
 Doctor’s Data has optimized identification of gastrointestinal microorganisms from cultured stool specimens using an accurate and reliable proteomic technique (MaLDI- TOF MS) that identifies bacteria and yeast by spectral analysis of microorganisms’ predominant ribosomal proteins.
Doctor’s Data has optimized identification of gastrointestinal microorganisms from cultured stool specimens using an accurate and reliable proteomic technique (MaLDI- TOF MS) that identifies bacteria and yeast by spectral analysis of microorganisms’ predominant ribosomal proteins.
PCR platforms, albeit less sensitive than culture, have a place in acute care situations of infectious diarrheal disease when time is of the essence. However the PCR approach does not currently lend itself to comprehensive analysis of the vast array of gastrointestinal microorganisms.
Currently, culture/MALDI- TOF MS technology is the most sensitive and discriminating way to identify the greatest number of beneficial/expected, commensal and pathogenic gastrointestinal microorganisms present in your patients.
Share:
Related Posts

TUDCA vs. Milk Thistle: Which One Supports Your Liver Best?
Written by Ashley Palmer & Pooja Mahtani | 2025 Key Takeaways: TUDCA is a bile acid that enhances bile flow, reduces liver inflammation, and supports

14 Common Questions About PectaSol Modified Citrus Pectin
Written by ecoNugenics An increased interest in natural therapies to treat the body has led to exciting research into a variety of supplements. One common

Benefits of Creatine in Perimenopause and Menopause
Written by Maura MacDonald, MS, RD, CSSD | 2025 As we age, the notion is that we will inevitably become weaker. Not as mobile as

Goodbye Pie Chart, Hello Phase 1 Sliders
Written by Allison Smith, ND | 2025 As we usher in a new era of DUTCH testing which leaves behind the concept of the three-way

Introducing the DUTCH Dozen
Written by Kelly Ruef, ND | 2025 Hormone testing can be complex, which is why Precision Analytical developed the DUTCH Dozen, an interpretive framework that


